# K-means Clustering
import numpy as np
import tensorflow.compat.v1 as tf
# unknown reason
from scipy.spatial import cKDTree
# in order to plot four-dimension data into two-dimension plot
from sklearn.decomposition import PCA
# import iris datasets
from sklearn import datasets
from sklearn.preprocessing import scale
# plot tools
import matplotlib.pyplot as plt
import pandas as pd
/opt/anaconda3/lib/python3.8/site-packages/requests/__init__.py:89: RequestsDependencyWarning: urllib3 (1.26.8) or chardet (3.0.4) doesn't match a supported version!
warnings.warn("urllib3 ({}) or chardet ({}) doesn't match a supported "
tf.disable_eager_execution()
tf.disable_v2_behavior()
WARNING:tensorflow:From /opt/anaconda3/lib/python3.8/site-packages/tensorflow/python/compat/v2_compat.py:111: disable_resource_variables (from tensorflow.python.ops.variable_scope) is deprecated and will be removed in a future version.
Instructions for updating:
non-resource variables are not supported in the long term
iris = datasets.load_iris()
iris_data = pd.DataFrame(data = iris.data)
iris_data
| 0 | 1 | 2 | 3 | |
|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 |
| ... | ... | ... | ... | ... |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 |
150 rows × 4 columns
num_pts = len(iris.data)
num_feats = len(iris.data[0])
sess = tf.Session()
num_pts, num_feats # number of pieces, number of features
(150, 4)
k = 3
generations = 25
data_points = tf.Variable(iris.data)
cluster_labels = tf.Variable(tf.zeros([num_pts],dtype=tf.int64))
rand_starts = np.array([iris.data[np.random.choice(len(iris.data))] for _ in range(k)])
rand_starts
array([[5. , 3.3, 1.4, 0.2],
[5.5, 2.3, 4. , 1.3],
[6.1, 2.8, 4. , 1.3]])
iris.data[1]
array([4.9, 3. , 1.4, 0.2])
centriods = tf.Variable(rand_starts)
centriods
<tf.Variable 'Variable_2:0' shape=(3, 4) dtype=float64_ref>
k, num_feats # centriods has 3 rows * 4 columns
(3, 4)
init = tf.initialize_all_variables()
sess.run(init)
WARNING:tensorflow:From /opt/anaconda3/lib/python3.8/site-packages/tensorflow/python/util/tf_should_use.py:247: initialize_all_variables (from tensorflow.python.ops.variables) is deprecated and will be removed after 2017-03-02. Instructions for updating: Use tf.global_variables_initializer instead.
centriod_matrix = tf.reshape(tf.tile(centriods, [num_pts, 1]), [num_pts, k, num_feats])
centriod_matrix
<tf.Tensor 'Reshape:0' shape=(150, 3, 4) dtype=float64>
centriods.shape # Dimension 3 represents the number of cluster and Dimension 4 represents the number of features.
TensorShape([Dimension(3), Dimension(4)])
point_matrix = tf.reshape(tf.tile(data_points, [1,k]),[num_pts,k, num_feats])
point_matrix
<tf.Tensor 'Reshape_1:0' shape=(150, 3, 4) dtype=float64>
data_points.shape # 150 is the number of sample pieces and 4 is the number of the features
TensorShape([Dimension(150), Dimension(4)])
tf.tile(data_points, [1,k]).shape
TensorShape([Dimension(150), Dimension(12)])
distances = tf.reduce_sum(tf.square(point_matrix-centriod_matrix), reduction_indices=2)
distances.shape
TensorShape([Dimension(150), Dimension(3)])
distances_1 = tf.square(point_matrix-centriod_matrix)
distances_1
<tf.Tensor 'Square_1:0' shape=(150, 3, 4) dtype=float64>
centriod_group = tf.argmin(distances,1) # why returning the index of the smallest distance between one particular point and three centriods?
centriod_group
tf.initialize_all_variables()
<tf.Operation 'init_1' type=NoOp>
centriod_group.shape
TensorShape([Dimension(150)])
sum_total = tf.unsorted_segment_sum(data_points,centriod_group,3)
num_total = tf.unsorted_segment_sum(tf.ones_like(data_points), centriod_group,3)
avg_by_group = sum_total/num_total
def data_group_avg(data, group_ids):
sum_total = tf.unsorted_segment_sum(data,group_ids,3)
num_total = tf.unsorted_segment_sum(tf.ones_like(data), group_ids,3)
avg_by_group = sum_total/num_total
return(avg_by_group)
means = data_group_avg(data_points,centriod_group)
update = tf.group(centriods.assign(means), cluster_labels.assign(centriod_group)) # tf.variable is very important function and should be not
# converted into the constants in tensorflow by using sess.run when you want to run for or while loops
for i in range(generations):
print('Calculating generation {}, out of {}'.format(i, generations))
_, centriod_group_count = sess.run([update, centriod_group])
group_count = []
for ix in range(k):
group_count.append(np.sum(centriod_group_count==ix))
print('Group counts: {}'.format(group_count))
Calculating generation 0, out of 25
Group counts: [50]
Group counts: [50, 18]
Group counts: [50, 18, 82]
Calculating generation 1, out of 25
Group counts: [50]
Group counts: [50, 31]
Group counts: [50, 31, 69]
Calculating generation 2, out of 25
Group counts: [50]
Group counts: [50, 36]
Group counts: [50, 36, 64]
Calculating generation 3, out of 25
Group counts: [50]
Group counts: [50, 40]
Group counts: [50, 40, 60]
Calculating generation 4, out of 25
Group counts: [50]
Group counts: [50, 45]
Group counts: [50, 45, 55]
Calculating generation 5, out of 25
Group counts: [50]
Group counts: [50, 49]
Group counts: [50, 49, 51]
Calculating generation 6, out of 25
Group counts: [50]
Group counts: [50, 54]
Group counts: [50, 54, 46]
Calculating generation 7, out of 25
Group counts: [50]
Group counts: [50, 57]
Group counts: [50, 57, 43]
Calculating generation 8, out of 25
Group counts: [50]
Group counts: [50, 60]
Group counts: [50, 60, 40]
Calculating generation 9, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 10, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 11, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 12, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 13, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 14, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 15, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 16, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 17, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 18, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 19, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 20, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 21, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 22, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 23, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
Calculating generation 24, out of 25
Group counts: [50]
Group counts: [50, 61]
Group counts: [50, 61, 39]
[centers, assignments] = sess.run([centriods,cluster_labels])
assignments
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 2, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 1, 2, 2, 2, 2, 1, 2, 2, 2,
2, 2, 2, 1, 1, 2, 2, 2, 2, 1, 2, 1, 2, 1, 2, 2, 1, 1, 2, 2, 2, 2,
2, 1, 2, 2, 2, 2, 1, 2, 2, 2, 1, 2, 2, 2, 1, 2, 2, 1])
max(set(list(assignments)[100:150]), key=list(assignments)[100:150].count)
2
[centers, assignments] = sess.run([centriods, cluster_labels])
def most_common(my_list):
return(max(set(my_list), key= my_list.count))
label0 = most_common(list(assignments)[0:50])
label1 = most_common(list(assignments)[50:100])
label2 = most_common(list(assignments)[100:150])
group0_count = np.sum(assignments[0:50]==label0)
group1_count = np.sum(assignments[50:100]==label1)
group2_count = np.sum(assignments[100:150]==label2)
print('Accuarcy: {}'.format((group0_count+group1_count+group2_count)/150))
Accuarcy: 0.8866666666666667
pca_model = PCA(n_components=2)
reduced_data = pca_model.fit_transform(iris.data)
reduced_centers = pca_model.transform(centers)
h = .02
x_min, x_max = reduced_data[:,0].min()-1, reduced_data[:,0].max()+1
y_min, y_max = reduced_data[:,1].min()-1, reduced_data[:,1].max()+1
xx, yy = np.meshgrid(np.arange(x_min,x_max,h),np.arange(y_min,y_max,h))
xx_pt = list(xx.ravel())
yy_pt = list(yy.ravel())
xy_pts = np.array([[x,y] for x, y in zip(xx_pt,yy_pt)])
mytree = cKDTree(reduced_centers)
dist, indexes = mytree.query(xy_pts)
indexes = indexes.reshape(xx.shape)
plt.clf()
plt.imshow(indexes, interpolation='nearest', extent=(xx.min(),xx.max(),yy.min(),yy.max()),
cmap = plt.cm.Paired,aspect = 'auto', origin = 'lower')
symbols = ['o', '^', 'D']
label_name = ['Setosa', 'Versicolour','Virginica']
for i in range(3):
temp_group = reduced_data[(i*50):(50)*(i+1)]
plt.plot(temp_group[:,0],temp_group[:,1],symbols[i], markersize = 10, label = label_name[i])
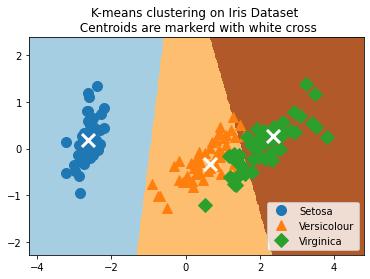
plt.scatter(reduced_centers[:, 0], reduced_centers[:,1], marker='x', s = 169, linewidths =3, color='w',zorder=10)
plt.title('K-means clustering on Iris Dataset \n Centroids are markerd with white cross')
plt.xlim(x_min, x_max)
plt.ylim(y_min, y_max)
plt.legend(loc='lower right')
plt.show()
K-Means clustering on the handwritten digits data¶
# load the dataset
import numpy as np
from sklearn.datasets import load_digits # digits datasets. Labels: 0,1,2,3,4,5,6,7,8,9. Features: 64 (grey image size: 8 width * 8 height).
# Sample size: 1797
data, labels = load_digits(return_X_y=True)
(n_samples, n_features), n_digits = data.shape, np.unique(labels).size
data
array([[ 0., 0., 5., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 10., 0., 0.],
[ 0., 0., 0., ..., 16., 9., 0.],
...,
[ 0., 0., 1., ..., 6., 0., 0.],
[ 0., 0., 2., ..., 12., 0., 0.],
[ 0., 0., 10., ..., 12., 1., 0.]])
labels
array([0, 1, 2, ..., 8, 9, 8])
n_samples, n_features
(1797, 64)
n_digits
10
print(f"# digits: {n_digits}; # samples: {n_samples}; # features: {n_features}")
# digits: 10; # samples: 1797; # features: 64
# kmeans = KMeans(init="kmeans++",n_cluster=n_digits, n_init=4, random_state=0)
from time import time # timing
from sklearn import metrics # metrics module for evaluation of performance: including score functions, performance metrics
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
def bench_k_means(kmeans, name, data, labels):
""" Benchmark to evaluate the KMeans initialzation methods.
Parameters
----------
kmeans: KMeans instance
A: class: '-sklearn.cluster.KMeans' instance with the initialization already set.
name: str
Name given to the strategy. It will be used to show the results in a table.
data: ndarray of shape (n_samples,)
The labels used to compute the clustering metrics which requires some supervision.
"""
t0 = time() # time clock starts
estimator = make_pipeline(StandardScaler(), kmeans).fit(data)
fit_time = time()-t0 # time clock ends. Fit-time is the time for the estimator
results = [name, fit_time, estimator[-1].inertia_]
# Define the metrics which require only the true labels and estimator labels
clustering_metrics = [
metrics.homogeneity_score,
metrics.completeness_score,
metrics.v_measure_score,
metrics.adjusted_rand_score,
metrics.adjusted_mutual_info_score,
]
results += [m(labels, estimator[-1].labels_) for m in clustering_metrics]
# The silhouette score requires the full dataset
results += [
metrics.silhouette_score(
data,
estimator[-1].labels_,
metric = "euclidean",
sample_size = 300,
)
]
# Show the results
formatter_result = (
"{:9s}\t{:.3f}s\t{:.0f}\t{:.3f}\t{:.3f}\t{:.3f}\t{:.3f}\t{:.3f}"
)
print(formatter_result.format(*results))
from sklearn.cluster import KMeans
from sklearn.decomposition import PCA
print(82*'-')
print("init\t\ttime\tinertia\thomo\tcompl\tv-meas\tARI\tAMI\tsilhouette")
kmeans = KMeans(init="k-means++", n_clusters = n_digits, n_init=4, random_state = 0)
bench_k_means(kmeans=kmeans, name= "k-means++", data = data, labels=labels)
kmeans = KMeans(init="random", n_clusters = n_digits, n_init=4, random_state = 0)
bench_k_means(kmeans=kmeans, name="random", data=data, labels=labels)
pca = PCA(n_components=n_digits).fit(data)
kmeans = KMeans(init=pca.components_,n_clusters=n_digits,n_init=1)
bench_k_means(kmeans=kmeans,name="PCA-based", data=data, labels=labels)
print(82*'-')
----------------------------------------------------------------------------------
init time inertia homo compl v-meas ARI AMI silhouette
k-means++ 0.126s 69485 0.613 0.660 0.636 0.482 0.632
random 0.043s 69952 0.545 0.616 0.578 0.415 0.574
PCA-based 0.019s 72686 0.636 0.658 0.647 0.521 0.643
----------------------------------------------------------------------------------
import matplotlib.pyplot as plt
reduced_data = PCA(n_components=2).fit_transform(data)
kmeans = KMeans(init="random", n_clusters=n_digits, n_init=4)
kmeans.fit(reduced_data)
h = 0.02
x_min, x_max = reduced_data[:,0].min()-1, reduced_data[:,0].max()+1
y_min, y_max = reduced_data[:,1].min()-1, reduced_data[:,1].max()+1
xx, yy = np.meshgrid(np.arange(x_min,x_max,h), np.arange(y_min,y_max,h))
Z = kmeans.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.figure()
plt.clf()
plt.figure(dpi=1000)
plt.imshow(
Z,
interpolation="nearest",
extent = (xx.min(), xx.max(), yy.min(), yy.max()),
cmap = plt.cm.Paired,
aspect = "auto",
origin= "lower",
)
plt.plot(reduced_data[:,0],reduced_data[:,1], 'k.', markersize=2)
centroids = kmeans.cluster_centers_
plt.scatter(
centroids[:,0],
centroids[:,1],
marker = "x",
s=169,
linewidths=3,
color = "w",
zorder = 10,
)
plt.title(
"K-means clustering on the digits dataset (PCA-reduced data) \n"
"Centroids are marked with white cross"
)
plt.xlim(x_min,x_max)
plt.ylim(y_min,y_max)
plt.xticks(())
plt.yticks(())
plt.show()
<Figure size 432x288 with 0 Axes>

K-Means Clustering of Iris Dataset¶
sess = tf.Session()
iris = datasets.load_iris()
num_pts = len(iris.data)
num_feats = len(iris.data[0])
k = 3
generations = 25
data_points = tf.Variable(iris.data)
cluster_labels = tf.Variable(tf.zeros([num_pts],dtype=tf.int64))
rand_starts = np.array([iris.data[np.random.choice(len(iris.data))] for _ in range(k)])
centriods = tf.Variable(rand_starts)
centriod_matrix = tf.reshape(tf.tile(centriods, [num_pts,1]), [num_pts, k, num_feats])
point_matrix = tf.reshape(tf.tile(data_points, [1,k]),[num_pts,k,num_feats])
distances = tf.reduce_sum(tf.square(point_matrix-centriod_matrix), reduction_indices=2)
centriod_group = tf.argmin(distances,1)
def data_group_avg(group_ids, data):
sum_total = tf.unsorted_segment_sum(data, group_ids,3)
num_total = tf.unsorted_segment_sum(tf.ones_like(data), group_ids,3)
avg_by_group = sum_total/num_total
return(avg_by_group)
means = data_group_avg(centriod_group, data_points)
update = tf.group(centriods.assign(means), cluster_labels.assign(centriod_group))
init = tf.initialize_all_variables()
sess.run(init)
for i in range(generations):
print('Calculating gen {}, out of {}'.format(i, generations))
_, centriod_group_count = sess.run([update, centriod_group])
group_count = []
for ix in range(k):
group_count.append(np.sum(centriod_group_count==ix))
print("Group counts: {}".format(group_count))
#[centers, assignments] = sess.run([centroids, cluster_labels])
def most_common(my_list):
return(max(set(my_list), key = my_list.count))
label0 = most_common(list(assignments[0:50]))
label1 = most_common(list(assignments[50:100]))
label2 = most_common(list(assignments[100:150]))
group0_count = np.sum(assignments[0:50]==label0)
group1_count = np.sum(assignments[50:100]==label1)
group2_count = np.sum(assignments[100:150]==label2)
accuracy = (group0_count+group1_count+group2_count)/150.
print('Accuracy: {:.2f}'.format(accuracy))
Calculating gen 0, out of 25
Group counts: [21, 79, 50]
Calculating gen 1, out of 25
Group counts: [31, 69, 50]
Calculating gen 2, out of 25
Group counts: [36, 64, 50]
Calculating gen 3, out of 25
Group counts: [40, 60, 50]
Calculating gen 4, out of 25
Group counts: [45, 55, 50]
Calculating gen 5, out of 25
Group counts: [49, 51, 50]
Calculating gen 6, out of 25
Group counts: [54, 46, 50]
Calculating gen 7, out of 25
Group counts: [57, 43, 50]
Calculating gen 8, out of 25
Group counts: [60, 40, 50]
Calculating gen 9, out of 25
Group counts: [61, 39, 50]
Calculating gen 10, out of 25
Group counts: [61, 39, 50]
Calculating gen 11, out of 25
Group counts: [61, 39, 50]
Calculating gen 12, out of 25
Group counts: [61, 39, 50]
Calculating gen 13, out of 25
Group counts: [61, 39, 50]
Calculating gen 14, out of 25
Group counts: [61, 39, 50]
Calculating gen 15, out of 25
Group counts: [61, 39, 50]
Calculating gen 16, out of 25
Group counts: [61, 39, 50]
Calculating gen 17, out of 25
Group counts: [61, 39, 50]
Calculating gen 18, out of 25
Group counts: [61, 39, 50]
Calculating gen 19, out of 25
Group counts: [61, 39, 50]
Calculating gen 20, out of 25
Group counts: [61, 39, 50]
Calculating gen 21, out of 25
Group counts: [61, 39, 50]
Calculating gen 22, out of 25
Group counts: [61, 39, 50]
Calculating gen 23, out of 25
Group counts: [61, 39, 50]
Calculating gen 24, out of 25
Group counts: [61, 39, 50]
Accuracy: 0.89
# To visually see our groupings and if they have indeed separated out the iris species, we will transform the four dimensions to
# two dimensions using PCA, and plot the data points and groups. After the PCA decomposition, we create predictions on a grid of
# x-y values for plotting a color graph:
pca_model = PCA(n_components=2)
reduced_data = pca_model.fit_transform(iris.data)
iris.data.shape, reduced_data.shape
((150, 4), (150, 2))
centers
array([[5.006 , 3.428 , 1.462 , 0.246 ],
[5.88360656, 2.74098361, 4.38852459, 1.43442623],
[6.85384615, 3.07692308, 5.71538462, 2.05384615]])
# Transform centers
reduced_centers = pca_model.transform(centers)
reduced_centers
array([[-2.64241546, 0.19088505],
[ 0.66567601, -0.3316042 ],
[ 2.34652659, 0.27393856]])